The genome of *Mediocremonas mediterraneus*
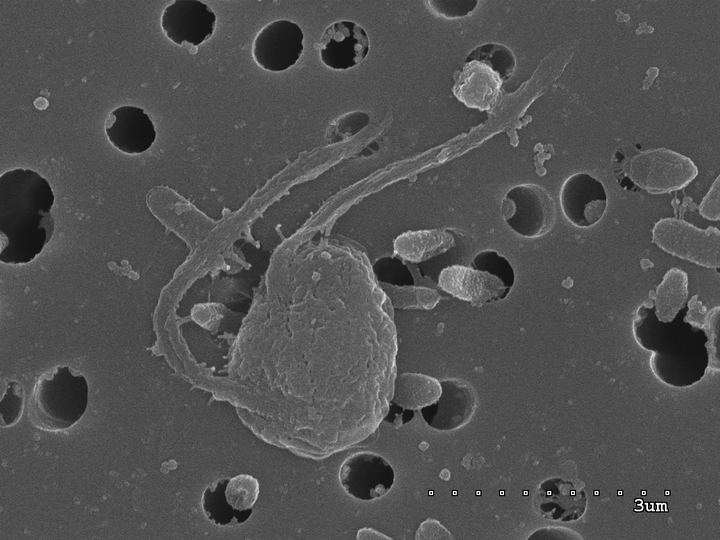 Photo by Javier del Campo
Photo by Javier del Campo
There is a significant bias in eukaryotic genomics that impoverishes our understanding of the diversity of eukaryotes, and leads to skewed views of what a eukaryote even is, as well as their role in the environment. This bias is simple and widely recognized: most genomics focuses on multicellular eukaryotes. Around 85% of the eukaryotic genomes correspond to multicellular organisms, either Metazoa, Fungi, or Land Plants. However, these lineages only represent 62% of the 18S rDNA GenBank sequences, which is a biased sample anyway, or 23% of all operational taxonomic units (OTUs) in environmental surveys. This makes for a pitiful future if we aim to understand and appreciate the whole eukaryotic tree of life. We need to change this trend, or we are going to neglect most eukaryotic diversity in the future genomic or metagenomic-based ecological and evolutionary studies. In this project we will generate the genome of Mediocremonas mediterraneus (Blanes Bay, Catalonia),a heterotrophic nanoflagellate belonging to the Developea within the supergroup Stramenopiles. Based on phylogenomics the Developea are sister to all the photosynthetic Stramenopiles (Diatoms, Kelps, etc.). The phylogenetic position of M. mediterraneus s makes it a good candidate to study the rise of photosynthesis in Stramenopiles.
Chytridiomycosis has already caused the disappearance of more than 200 species of amphibians around the planet. These diseases, very virulent and with very harmful effects on amphibians, are believed to have been exacerbated by the effects of climate change, globalization, pollution and, above all, by the effect of exotic amphibian species introduced through trade. of legal and illegal species and that act as transmission vectors. Due to all these threats, many amphibian species and populations have been predicted to disappear in the coming years. Of all the amphibian species, it has been seen that those with a higher risk of extinction are mainly those with smaller distributions and fewer populations in mountainous areas that are being transformed as a result of human activity. This situation of population reduction and loss of quantity and quality of habitat, makes these amphibian populations much more susceptible to other threats, including climate change and, above all, infectious diseases caused by chytrid fungi. Calotriton arnoldi is the only European salamander considered Critically Endangered by the IUCN, a category that makes it the most threatened amphibian in Europe. Calotriton arnoldi was described in 2005 from populations that inhabit the Montseny Natural Park (Catalonia). The only known populations of Calotriton arnoldi correspond to three mountain streams in the eastern part and four streams in the western part of the Tordera River valley. It has been calculated that the occupation area of C. arnoldi is only 10 km2. Another recent threat linked to climate change that also stalks the Montseny newt are the already mentioned emerging diseases caused by chytrid fungi. Although to date these diseases have not been detected in C. arnoldi populations, recent studies have confirmed an outbreak of the chytrids Bd and especially Bsal in the Montnegre i el Corredor Natural Park, just 15 Km South of the Montseny massif. Our goal in this project is to find bacterial candidates from C. arnoldi’s skin microbiome that could be used as a probiotic under a similar bioaugmentation strategy to fight Bsal. We will compare the microbiomes of C. arnoldi with those from T. marmoratus and S. salamandra individuals that appear to be correlated to resistance against disease. Based on the results obtained from this comparison we will recover the frozen samples from C. arnoldi individuals that present microbiomes more similar to those resistant newts and salamanders from the Montnegre i el Corredor. We will use the samples to isolate bacteria in the laboratory in order to create a collection of putative probiotics from the newt’s skin microbiota with potential antifungal activity. We will do that in parallel on T. marmoratus and S. salamander samples from resistant individuals. This collection of bacterial isolates will be screened in the lab by testing its antifungal activity against Bsal cultures.